Microbiomes Move Standard Plate Count One Step Forward
This article is part 2 of the Food Genomics column published by Food Safety Tech. Read the original article here.
These microorganisms open the door to the vast and varied biosphere of the microbiology of food.
Last month we introduced several food genomics terms including the microbiome. Recall that a microbiome is the community or population of microorganisms that inhabit a particular environment or sample. Recall that there are two broad types of microbiomes, a targeted (e.g., bacteria or fungi) or a metagenome (in which all DNA in a sample is sequenced, not just specific targets like bacteria or fungi). This month we would like to introduce the reader to uses of microbiomes and how they augment standard plate counts and move us into a new era in food microbiology. Before providing examples, it might be useful to review a diagram explaining the general flow of the process of determining a microbiome (See Figure 1).
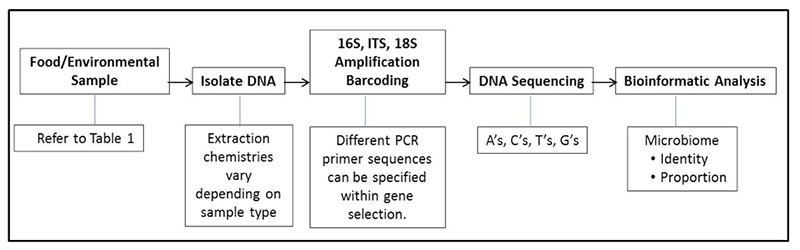 Figure 1. General process for performing a targeted microbiome (bacterial or fungal)
Figure 1. General process for performing a targeted microbiome (bacterial or fungal)
By analogy, if one thinks of cultural microbiology and plate counts as a process of counting colonies of microbes that come from a food or environmental sample, microbiome analysis can be thought of as identifying and counting signature genes, such as the bacterial specific 16S gene, from the microbes in a food or environmental sample. Plate counts have been and remain a food microbiologist most powerful indicator tool in the tool kit; however, we know there are some limitations in their use. One limitation is that not all bacterial or fungal cells are capable of outgrowth and colony formation on specific media under a set of incubation conditions (temperature, time, media pH, storage atmosphere, etc.). Individual plate count methods cannot cover the nearly infinite number of variations of growth atmospheres and nutrients. Because of these limitations microbiologists understand that we have not cultured but many different types of bacteria on the planet (this led to the term “The Great Plate Count Anomaly” (Staley & Konopka, 1985). Think of a holiday party where guests were handed nametags on which was printed: “Hello, I grow on Standard Methods Agar” or “Hello, I grow at 15°C”, etc. We can group the partygoers by ability to grow on certain media; we can also count partygoers, but they still do not have names. As effective as our selective and differential media have become, bacterial colonies still do not come with their own “Hello, My Name Is XYZ” name tags. Therefore, in the lab, once a plate is counted it is generally tossed into the autoclave bag, along with unnamed colonies and all they represent. Microbiomes can provide a nametag of sorts as well as what proportion of people at that party share a certain name. For instance: “Hello, My Name is Pseudomonas” or “Hello, My Name is Lactobacillus”, etc. The host can then say “Now we are going to count you; would all Pseudomonas pleased gather in this corner?” or “All Lactobacillus please meet at the punch bowl”.
A somewhat overly simplified analogy, but it makes the point that microbiome technology gives names and proportions. Microbiomes too have limitations. First, with current technologies microbiomes need a relatively large threshold of organisms of a specific group to appear in the microbiome pie chart— approximately 103. In theory, a colony on a plate of agar medium can be derived from a single cell or colony-forming unit (CFU). Not all amplified genes in a microbiome are necessarily from viable cells (A topic that will be covered later in this series of articles). Forming a colony on an agar surface on the other hand requires cell viability. Finally, the specificity of a microorganism name assigned to a group in a microbiome depends on the size of the sequenced amplicon (an amplicon is a segment of DNA, in this case the 16S gene DNA, resulting from amplification by PCR before sequencing) and how well our microbial databases cover different subtypes in a species. Targeted microbiomes can reliably name the genus of an organism, however resolution to the species and subspecies is not guaranteed. (Later in this series we will discuss metagenomes and how they have the potential to identify to a species or even subspecies level). Readers can find very informative reviews on microbiome specificity in the following cited references: Bokulich, Lewis, Boundy-Mills, & Mills, 2016; de Boer et al., 2015; Ercolini, 2013; Kergourlay, Taminiau, Daube, & Champomier Vergès, 2015.
When we consider the power of using cultural microbiology for quantitative functional indicators of microbial quality together with microbiomic analysis, with limitations and all for both, microbiomes have opened a door to the vast and varied biosphere of our food’s microbiology to a depth never before observed. This all sounds great, but how will we benefit and use this information? We have constructed Table 1 with examples and links of microbiome applications to problems that would have required years to study by cultural microbiology techniques alone. Please note this is by no means an exhaustive list, but it serves to illustrate the very broad and deep potential of microbiomics to food microbiology. We encourage the reader to email the editors or authors with questions regarding any reference. Using PubMed and the search terms “Food AND microbiome” will provide abstracts and a large variety of applications of this technology.
| Foodstuff | Reference |
| Ale | (Bokulich, Bamforth, & Mills, 2012) |
| Beef Burgers | (Ferrocino et al., 2015) |
| Beefsteak | (De Filippis, La Storia, Villani, & Ercolini, 2013) |
| Brewhouse and Ingredients | (Bokulich et al., 2012) |
| Cheese | (Wolfe, Button, Santarelli, & Dutton, 2014) |
| Cheese and Listeria growth | (Callon, Retureau, Didienne, & Montel, 2014) |
| Cherries, Hydrostatic Pressure | (del Árbol et al., n.d.) |
| Cocoa | (Illeghems, De Vuyst, Papalexandratou, & Weckx, 2012) |
| Dairy Starters and Spoilage Bacteria | (Stellato, De Filippis, La Storia, & Ercolini, 2015) |
| Drinking Water Biofilms | (Chao, Mao, Wang, & Zhang, 2015) |
| Fermented Foods | (Tamang, Watanabe, & Holzapfel, 2016) |
| Foodservice Surfaces | (Stellato, La Storia, Cirillo, & Ercolini, 2015) |
| Fruit and Vegetables | (Leff & Fierer, 2013) |
| Insect Protein | (Garofalo et al., 2017) |
| Kitchen surfaces | (Flores et al., 2013) |
| Lamb | (Wang et al., 2016) |
| Lobster | (Tirloni, Stella, Gennari, Colombo, & Bernardi, 2016) |
| Meat and storage atmosphere | (Säde, Penttinen, Björkroth, & Hultman, 2017) |
| Meat spoilage and processing plant | (Pothakos, Stellato, Ercolini, & Devlieghere, 2015) |
| Meat Spoilage Volatiles | (Casaburi, Piombino, Nychas, Villani, & Ercolini, 2015) |
| Meat Stored in Different Atmospheres | (Ercolini et al., 2011) |
| Milk | (Quigley et al., 2011) |
| Milk and Cow Diet | (Giello et al., n.d.) |
| Milk and Mastitis | (Bhatt et al., 2012) |
| Milk and Teat Preparation | (Doyle, Gleeson, O’Toole, & Cotter, 2016) |
| Natural starter cultures | (Parente et al., 2016) |
| Olives | (Abriouel, Benomar, Lucas, & Gálvez, 2011) |
| Pork Sausage | (Benson et al., 2014) |
| Spores in complex foods | (de Boer et al., 2015) |
| Tomato Plants | (Ottesen et al., 2013) |
| Winemaking | (Marzano et al., 2016) |
| Table 1. Examples of microbiome analysis of different foods and surfaces. | |
See page 2 for references
This news piece was originally written by Gregory Siragusa, Senior Principal Scientist. News piece is posted on FoodSafety Tech.
